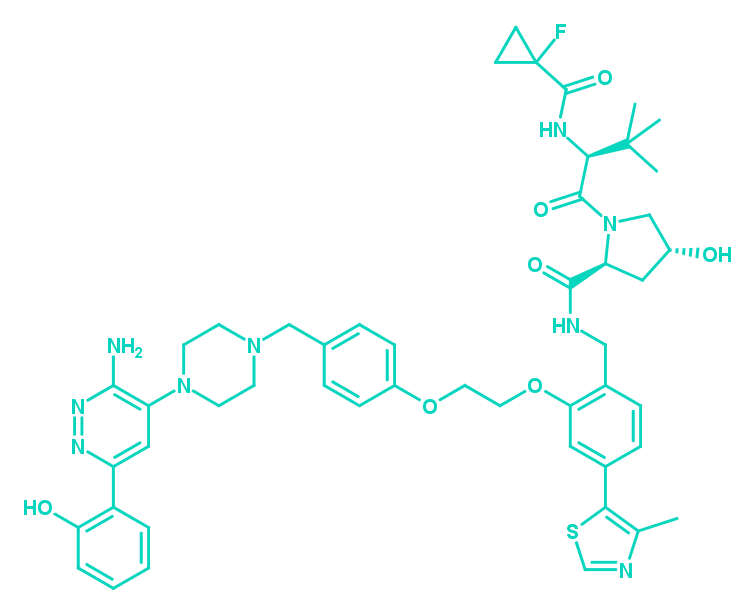
SMARCA2/4 PROTAC | ACBI1
Highlights
ACBI1 is a potent and cooperative PROTAC degrader of the BAF complex subunits SMARCA2, SMARCA4 and PRBM1. It harnesses the VHL E3 ligase to recruit its targets via their bromodomains, which results in their selective and pronounced degradation. This compound is suitable for both in vitro and in vivo studies and may be a useful tool to study the effects of an acute inactivation of the BAF complex.



Background information
Target Information
Would you like to learn more about the role of SMARCA2/4 in a new potential disease indication?
Watch the recording of a recent online seminar now.
The ATP-dependent activities of the BAF (SWI/SNF) chromatin remodeling complexes affect the positioning of nucleosomes on DNA and thereby many cellular processes related to chromatin structure, including transcription and DNA repair. The SMARCA2 and SMARCA4 genes code for the two alternative catalytic ATP-dependent helicases of the complex. Subunits of the BAF complexes are mutated in approximately 20% of human cancers. Several types of cells have been shown to be vulnerable to the loss of BAF complex ATPase subunits and subunits of the BAF complexes have been found to be mutated in approximately 20% of human cancers. Acute myeloid leukemia cells are sensitive to the loss of SMARCA4, whereas SMARCA4 mutant cancer cells are sensitive to the loss of SMARCA2. Future studies are warranted towards deeper mechanistic understanding of the downstream effects of degrading BAF ATPase subunits in both cancerous and non-cancerous cells. We hope that progress in this direction will be greatly advanced by sharing the chemical tool that has emerged from our work with the wider community.
ACBI1 is a PROTAC* that causes degradation of SMARCA2 and SMARCA4, as well as the facultative BAF complex subunit PBRM1. It is useful to study the consequences of inactivation of the BAF complex in cells in vitro, such as effects on chromatin accessibility, transcription and proliferation1-5 and can be also used for in vivo studies.
The compound is the result of a collaboration between the University of Dundee, School of Life Sciences, and Boehringer Ingelheim.
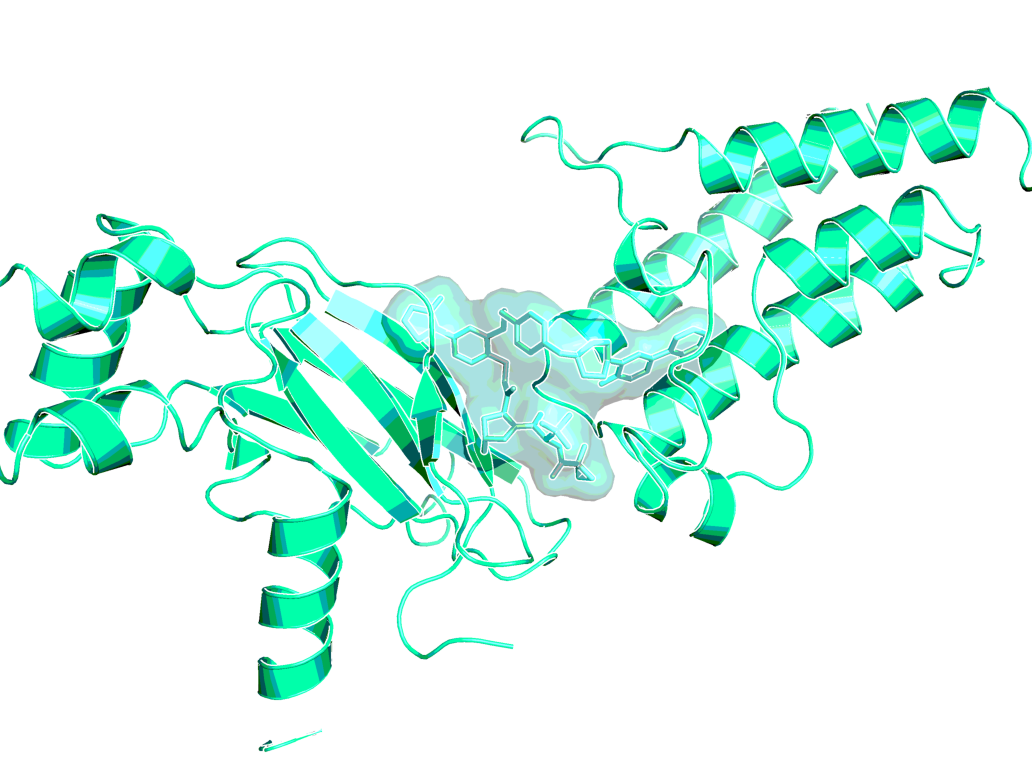
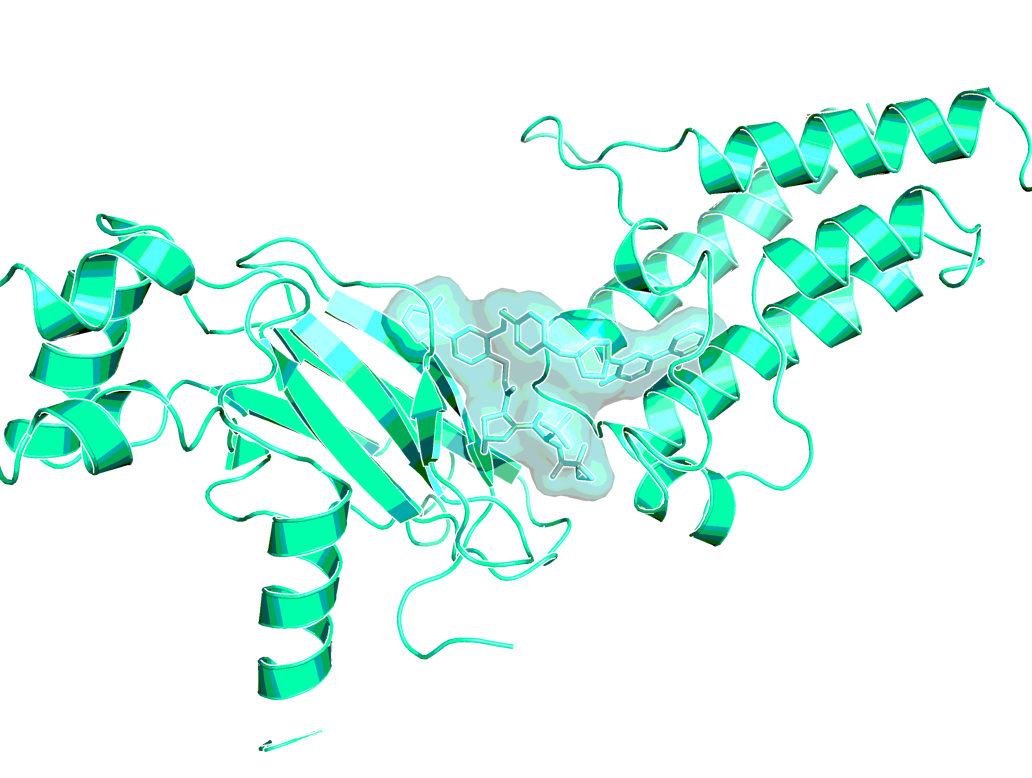
ACBI1 analog in ternary co-crystal structure of SMARCA2BD:ACBI1analog:VCB (PDB ID 6HAX)
*PROTACs (proteolysis-targeting chimeras) offer a novel drug modality wherein bi-functional molecules recruit a target protein and an E3 ligase to induce ubiquitylation and subsequently proteasomal degradation of the target protein.
In vitro activity
ACBI1 causes degradation of its targets in tissue culture cells.
| Probe name / negative control | ACBI1 | cis-ACBI1 |
| MW [Da] | 936.1 | 936.1 |
| Degradation of SMARCA2 in MV-4-11 cells (IC50) [nM]a | 6 | >1,000 |
| Degradation of SMARCA4 in MV-4-11 cells (IC50) [nM]a | 11 | >1,000 |
| Degradation of PBRM1 in MV-4-11 cells (IC50) [nM]a | 32 | >1,000 |
| Proliferation assay in MV-4-11 cells (IC50) [nM]b | 29 | 1400 |
| Proliferation assay in SK-MEL-5 cells (IC50) [nM]b | 77 | >10,000 |
a Treatment for 18h followed by analysis by capillary electrophoresis.
b Treatment for 7 days before measuring cell viability via cellular ATP content.
In vitro DMPK and CMC parameters
| Probe name / negative control | ACBI1 | cis-ACBI1 |
| Solubility @ pH 6.8 [µg/ml] | <1 | <1 |
| CACO permeability Pa-b @ pH7.4 [*10-6 cm/s] | 2.2 | n.d. |
| CACO efflux ratio | 1.7 | n.d. |
| Microsomal stability (human/mouse/rat) [% QH] | 86 / 46 / <23 | >88 / 53 / 34 |
| Hepatocyte stability* (human/mouse/rat) [% QH] | -/62/41 | n.d. |
| Plasma protein binding (FCS 10%) [%] | >99.4 | >89.2 |
| CYP 3A4 (IC50) [µM] | 0.4 | n.d. |
| CYP 2C8 (IC50) [µM] | <0.2 | n.d. |
| CYP 2C9 (IC50) [µM] | >50 | n.d. |
| CYP 2C19 (IC50) [µM] | 1.3 | n.d. |
| CYP 2D6 (IC50) [µM] | >50 | n.d. |
* in 50% plasma
In vivo DMPK parameters
ACBI1 | MOUSE | RAT |
| Clearance [% QH]a | 7.3 | 93 |
| Mean residence time [h]a | 0.8 | 2.8 |
| tmax [h]b | 0.33 | 1.5 |
| F [%]b | Quantitatively bioavailable | Quantitatively bioavailable |
| Vss [L/kg]a | 0.32 | 12 |
| AUC0-∞ [h nmol L-1]a | 14,000 | 1,500 |
| AUC0-∞ [h nmol L-1]b | 18,000 | 1,200 |
| AUD dose-normalized [h kg L-1]a | 2.6 | 0.29 |
| AUD dose-normalized [h kg L-1]b | 3.3 | 0.23 |
a 5 mg/kg i.v.; b 5 mg/kg s.c.
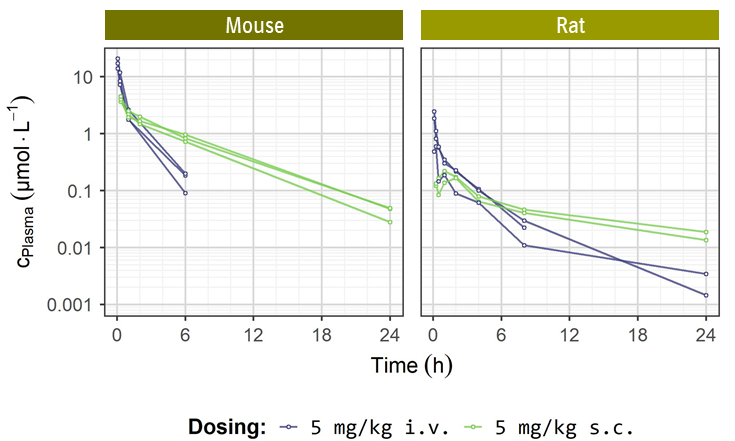
PK profiles of ACBI1 upon 5 mg/kg i.v. (blue curves) and 5 mg/kg s.c. dosing (green curves) in mouse and rat.
Negative control
In the negative control compound, cis-ACBI1, the hydroxy proline of the VHL binding part is in the inactive cis-conformation, which prevents binding to VHL and thereby protein degradation. This prevents binding to VHL, formation of a triple complex and thereby protein degradation. This compound is useful to differentiate phenotypic effects not caused by protein degradation in comparison with its active trans isomer ACBI1. Furthermore, the difference between binding to the bromodomain of SMARCA2 and SMARCA4 and its degradation can be studied in vitro.

Structure of cis-ACBI1 which serves as a negative control
Selectivity
The PROTAC ACBI1 causes selective degradation of SMARCA2, SMARCA4 and PBRM1.
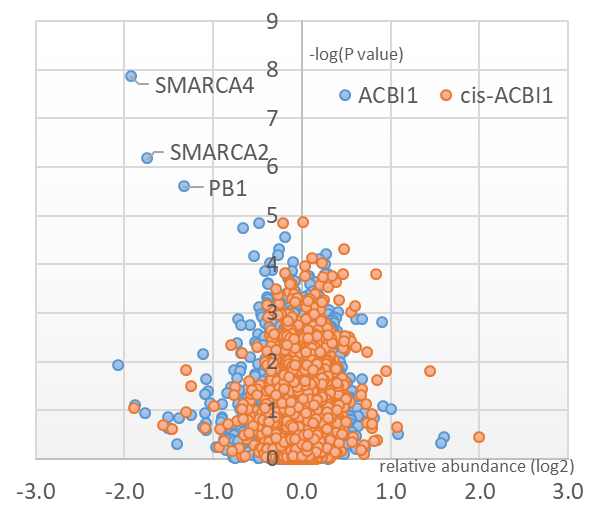
Multiplexed isobaric tagging mass spectrometry was utilised to demonstrate whole proteome selectivity of ACBI1 induced degradation.
Data show that amongst 6586 quantified proteins, only SMARCA2, SMARCA4 and PBRM1 are degraded. Of note, the levels of other BAF sub-units including BCL7A and ACTL6A remain unchanged.1
| SELECTIVITY DATA AVAILABLE | ACBI1 | cis-ACBI1 |
SafetyScreen44™ with kind support of  | Yes | Yes |
| Invitrogen® | No | No |
| DiscoverX® | No | No |
| Dundee | No | No |
The PROTAC ACBI1 was tested by the Eric Fisher Laboratory - Dana-Farber Cancer Institute as part of their Degradation Proteomics Initiative.6,7 It induces selective degradation of SMARCA2 and SMARCA4 after 5 h of treatment at 1 µM in the neuroblastoma cell line Kelly Cells.
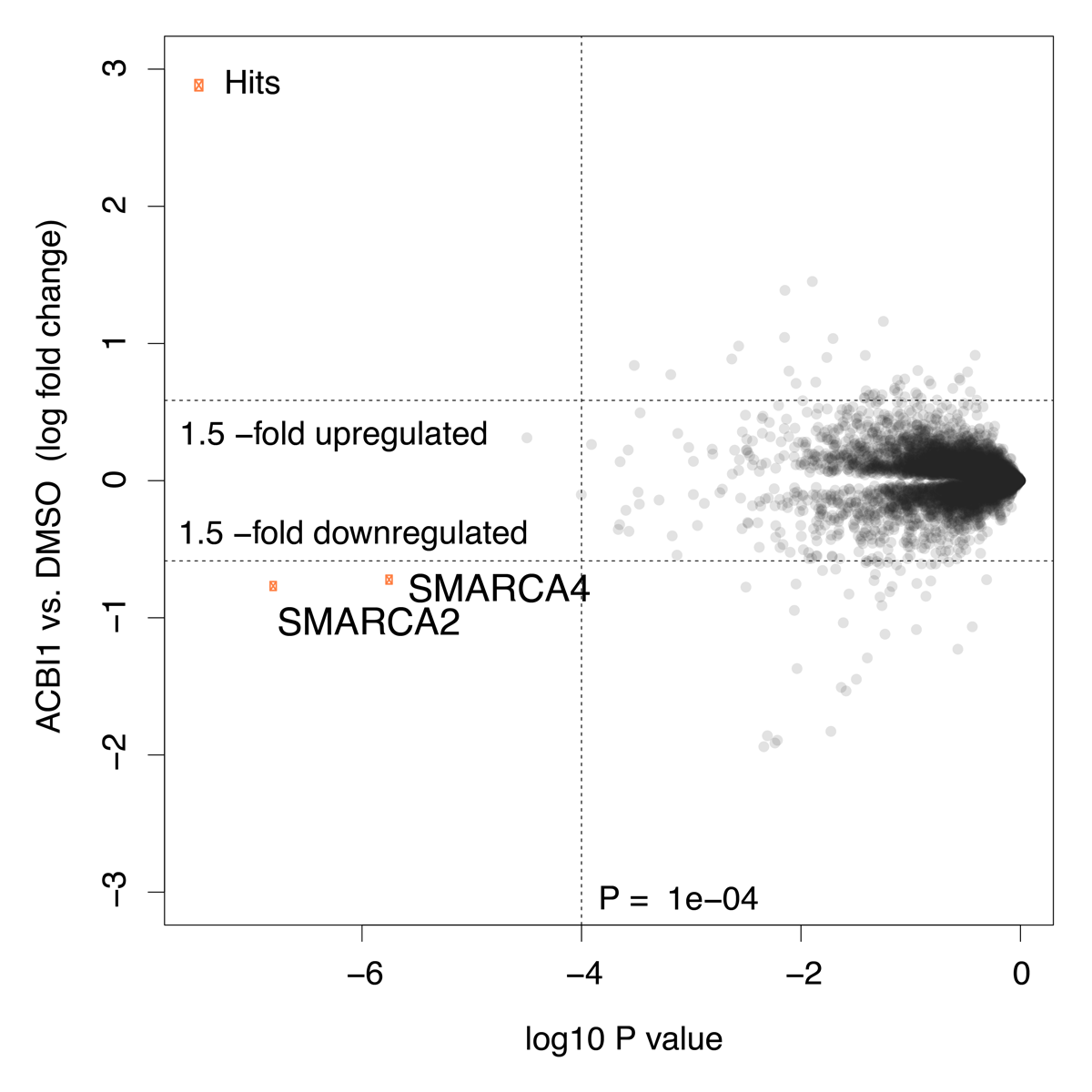
Global protein quantification was used to explore the unbiased proteome-wide selectivity of ACBI1 induced degradation. Whole cell protein quantification was performed using label free quantification with the Fischer lab’s diaPASEF workflow.
Of the 7,742 proteins quantified in this experiment, only SMARCA2 and SMARCA4 were found to be significantly downregulated in response to ACBI1 treatment. Statistical analysis was performed using a moderated t-test in Bioconductor’s limma package to generate hit lists containing log2 Fold Change and P-values for each protein. The data are also displayed in the scatterplot above.
Download selectivity data:
ACBI1_selectivityData.xlsx
ACBI1_neg_ctrl_selectivityData.xlsx
Degradation_Proteomics_Initiative_data_ACBI1-SMARCA-2-4-PROTAC_2.xlsx
Summary
ACBI1 causes pronounced degradation of the BAF chromatin remodeling complex ATPase proteins SMARCA2 and SMARCA4.
Supplementary data
2 D structure formats available
SMARCA2/4 PROTAC | ACBI1.smiles
Negative control | SMARCA2/4 PROTAC.png
References
BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design
Farnaby W.; Koegl M.; Roy M. J.; Whitworth C.; Diers E.; Trainor N.; Zollman D.; Steurer S.; Karolyi-Oezguer J.; Riedmueller C.; Gmaschitz T.; Wachter J.; Dank C.; Galant M.; Sharps B.; Rumpel K.; Traxler E.; Gerstberger T.; Schnitzer R.; Petermann O.; Greb P.; Weinstabl H.; Bader G.; Zoephel A.; Weiss-Puxbaum A.; Ehrenhöfer-Wölfer K.; Wöhrle S.; Boehmelt G.; Rinnenthal J.; Arnhof H.; Wiechens N.; Wu MY.; Owen-Hughes T.; Ettmayer P.; Pearson M.; McConnell D.B. and Ciulli A.
Nat Chem Biol. 2019, 15(7):672-680.
The SMARCA2/4 ATPase Domain Surpasses the Bromodomain as a Drug Target in SWI/SNF-Mutant Cancers: Insights from cDNA Rescue and PFI-3 Inhibitor Studies
Vangamudi B.; Paul T. A.; Shah P. K.; Kost-Alimova M.; Nottebaum L.; Shi X.; Zhan Y.; Leo E.; Mahadeshwar H. S.; Protopopov A.; Futreal A.; Tieu T. N.; Peoples M.; Heffernan T. P.; Marszalek J. R.; Toniatti C.; Petrocchi A.; Verhelle D.; Owen D. R.; Draetta G.; Jones P.; Palmer W. S.; Sharma S. and Andersen J. N.
Cancer Res. 2015, 75(18):3865-3878.
Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel–Lindau (VHL) E3 Ubiquitin Ligase: Structure–Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydr
Soares P.; Gadd M. S.; Frost J.; Galdeano C.; Ellis L.; Epemolu O.; Rocha S.; Read K. D.; Ciulli A.
J. Med. Chem. 2018, 61(2):599-618.
Mapping the Degradable Kinome Provides a Resource for Expedited Degrader Development
Donovan K. A., Ferguson F. M., Bushman J. W., Eleuteri N. A., Bhunia D., Ryu S., Tan L., Shi K., Yue H., Liu X., Dobrovolsky D., Jiang B., Wang J., Hao M., You I., Teng M., Liang Y., Hatcher J., Li Z., Manz T. D., Groendyke B., Hu W., Nam Y., Sengupta S., Cho H., Shin I., Agius M. P., Ghobrial I. M., Ma M. W., Che J., Buhrlage S. J., Sim T., Gray N. S., Fischer E. S.
Cell. 2020, 183(6):1714-1731.
How to cite opnMe?
When you plan a publication, please use the following acknowledgement:
ACBI1 was kindly provided by Boehringer Ingelheim via its open innovation platform opnMe, available at https://opnme.com.
Papers with our molecules. Published by you.
Acute BAF perturbation causes immediate changes in chromatin accessibility
Schick S., Grosche S., Kohl K. E., Drpic D., Jaeger M. G., Marella N. C., Imrichova H., Lin J-M. G., Hofstätter G., Schuster M., Rendeiro A. F., Koren A., Petronczki M., Bock C., Müller A. C., Winter G. E., Kubicek S.
Nat Genet 2021, 53(3):269-278.
BAF complexes drive proliferation and block myogenic differentiation in fusion-positive rhabdomyosarcoma
Laubscher D., Gryder B. E., Sunkel B. D., Andresson T., Wachtel M., Das S., Roschitzki B., Wolski W., Wu X. S., Chou H-C., Song Y. K., Wang C., Wei J. S., Wang M., Wen X., Ngo Q. A., Marques J. G., Vakoc C. R., Schäfer B. W., Stanton B. Z., Khan J.
Nat Commun 2021, 12(1):6924. open access
SMARCA4 biology in alveolar rhabdomyosarcoma
Bharathy N., Cleary M. M., Kim J-A., Nagamori K., Crawford K. A., Wang E., Saha D., Settelmeyer T. P., Purohit R., Skopelitis D., Chang K., Doran J. A., Kirschbaum C. W., Bharathy S., Crews D. W., Randolph M. E., Karnezis A. N., Hudson-Price L., Dhawan J., Michalek J. E., Ciulli A., Vakoc C. R., Keller C.
Oncogene 2022, 41(11):1647-1656.



