Illuminating the GPCRome
The opnMe GPCR Route 66+ project: Which of your fluorescent ligands can shed light on the selectivity of GPCR-targeting compounds to support faster drug discovery efforts?


Benedict-Tilman Berger, Felix Schiele
High Throughput Biology
Boehringer Ingelheim
subscription to the newsletter
As member of the GPCR community, join us for our new opnMe GPCR Route 66+ project. Contribute novel tracers to set up a defined GPCR selectivity binding assay panel based on the TR-FRET technology. Get reimbursed for shipment costs and receive exclusive access to all data that will be generated for your tracer(s). Win an award of 2,000 euros for each accepted and validated fluorescent tracer as part of Phase 2 of our call. Take the chance to co-author a scientific publication to improve early drug safety screening in a high impact factor journal upon conclusion of this call. Discover more…
Background information
What is the context of the problem that we would like to solve?
Cellular signaling pathways involving G protein-coupled receptors (GPCRs) regulate a wide range of physiological processes, such as vision, olfaction, hormone regulation, and neurotransmission, among others1,2. Consequently, GPCRs can play central roles in several pathologies, which make them prime targets of several successful medicines and many ongoing drug discovery efforts. Owing to their multiple functions, GPCRs can also be the source of drug adverse effects, such as emesis, sedation, and weight gain. Thus, by understanding GPCR selectivity of drug candidates, we can improve their therapeutic index that is a quantitative measurement of relative compound safety. With safety liabilities being the main cause of attrition in early drug development, the identification of potentially unsafe drugs in early phases of the discovery process can help to reduce the overall costs and timelines. To this end, fast and resource-effective methodologies to detect unselective GPCR binding can streamline the selection of safe candidates.
Currently, there are different assay systems commercially available that can be used to investigate GPCR modulation. Although assays such as beta-arrestin recruitment, calcium influx, receptor internalization, and second messenger production are commonly used in GPCR small molecule research, they primarily monitor the GPCR's function. However, these assays do not provide sufficient data resolution to determine a clear mechanism of action regarding the binding and kinetics of GPCR drugs. Therefore, binding characteristics such as kinetic selectivity – a parameter of increasing importance in pharmacometrics - cannot be thoroughly investigated.
To enable the scientific community to study the molecular mechanism of ligand – GPCRs interaction, scientists from the High Throughput Biology department at Boehringer Ingelheim have launched the opnMe GPCR Route 66+ project. The goal is to setup a GPCR selectivity binding assay panel based on the TR-FRET3 technology. This should enable large-scale profiling of the binding affinities and kinetics of test compounds for a variety of receptors. As a tracer displacement technology, TR-FRET may enable research on tracer based binding kinetics, molecular mechanism of action studies and determination of orthosteric or allosteric binding sites based on the tracer properties. To facilitate the process, we prepared 66 suitable GPCR constructs that are ready for human cell culture expression. To fulfill our mission of providing the scientific community with a high-quality, broad GPCR selectivity panel, we require your assistance in obtaining suitable fluorescent probes, also known as tracers, which are currently limited.
With the opnMe GPCR Route 66+ call, we are reaching out to the GPCR community to contribute novel tracers to expand the breadth of the panel to the highest possible number of targets. To this end, we offer our commitment to establish and validate assays using these tracers at our own costs, and to share the resulting protocol including documentation with the probe owners first, and with the scientific community later as part of an open access publication co-authored by all contributors of the final tracer set. In addition, we provide contributors of the final tracer set with the opportunity to propose candidate molecules for inclusion in the tests on the resulting broad GPCR panel to determine their affinities and kinetic binding properties.
This open science collaboration should rapidly enable scientists across the globe to perform comprehensive GPCR selectivity profiling for their compounds of interest, which will ultimately benefit the development of safer and more effective drugs.
What potential solutions could be in scope?
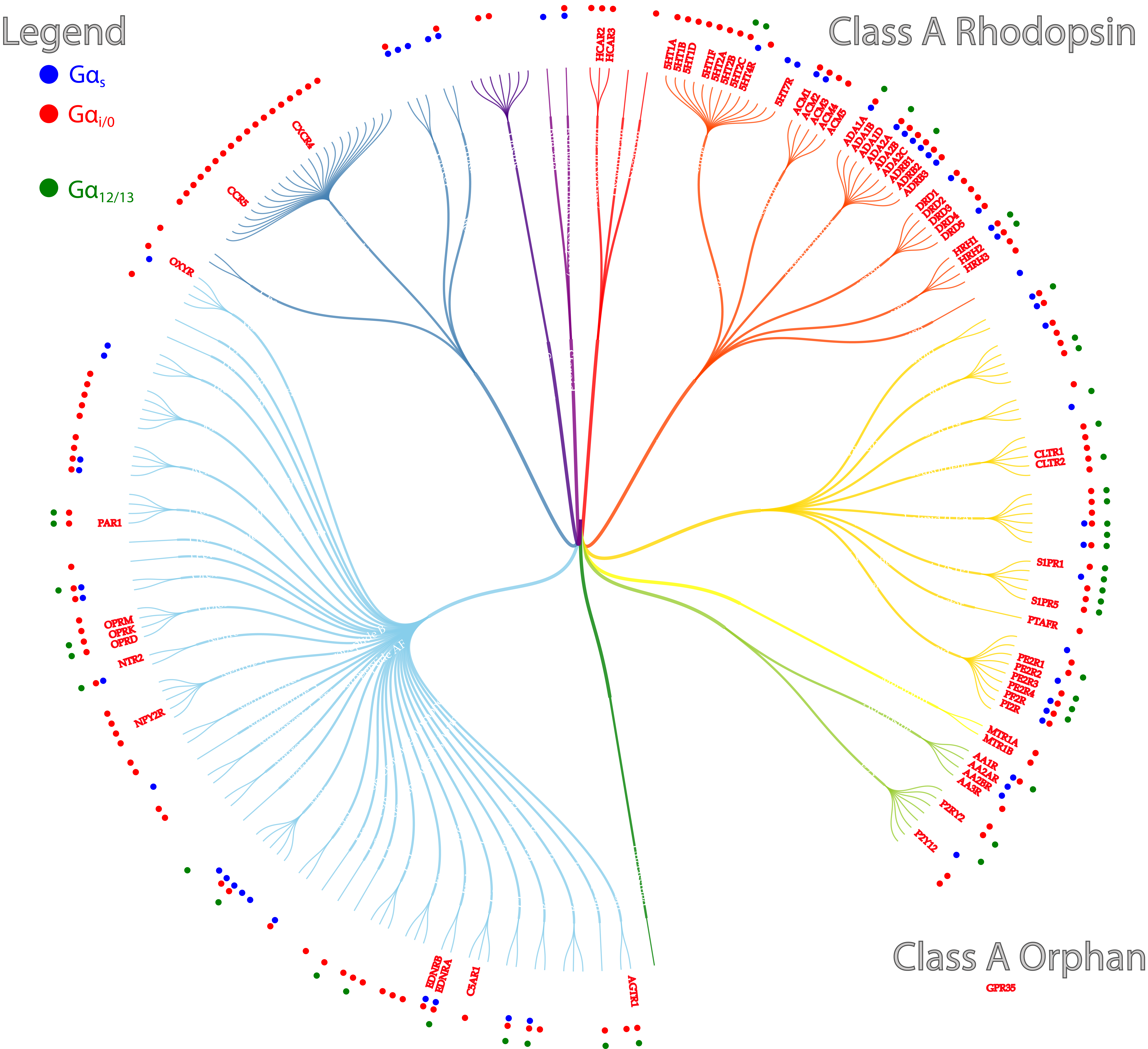
The scope of your contributions includes any fluorescently labelled ligand that binds ortho- or allosterically to one or more of the 66 GPCRs of interest for this project. Find an overview of the list and a depiction (figure 1) below and the complete list as part of the appendix (Excel file for download).:
Figure 1: Overview of the 66 GPCRs of interest in this call. GPCRs of interest are written in red. The signaling according to GPCRdb is shown as presented in the figure legend. Picture modified from reference 4.
|
GPCR subfamily/ligand |
GPCR of interest |
|
5-Hydroxytryptamine |
5HT1A, 5HT1B, 5HT1D, 5HT1F, 5HT2A, 5HT2B, 5HT2C, 5HT4R, 5HT7R |
|
Acetylcholine |
ACM1, ACM2, ACM3, ACM4, ACM5 |
|
Adenosine |
AA1R, AA2AR, AA2BR, AA3R |
|
Adrenaline |
ADA1A, ADA1B, ADA1D, ADA2A, ADA2B, ADA2C, ADRB1, ADRB2, ADRB3 |
|
Chemokine |
CCR5, CXCR4 |
|
Dopamine |
DRD1, DRD2, DRD3, DRD4, DRD5 |
|
Endothelin |
EDNRA, EDNRB |
|
Histamine |
HRH1, HRH2, HRH3 |
|
Hydroxycarboxylic acid |
HCAR2, HCAR3 |
|
Leukotriene |
CLTR1, CLTR2 |
|
Melatonine |
MTR1A, MTR1B |
|
Opioid |
OPRD, OPRK, OPRM |
|
P2Y |
P2RY2, P2Y12 |
|
Prostaglandin |
PE2R1, PE2R2, PE2R3, PE2R4, PI2R, PF2R |
|
S1P |
S1PR1, S1PR5 |
|
Other |
AGTR1, C5AR1, NPY2R, NTR2, OXYR, PTAFR, PAR1, GPR35 |
In addition, the profile of these ligands should fulfill the following criteria:
- They can be fragments, and/or small molecules, and/or peptides, and/or any biologics (antibodies, nanobodies, aptamers, etc.) and/or endogenous ligands with affinities (KD) for the considered GPCR better than 300 nM.
- Their fluorescent moieties should be excitable at 490, 548, 587 or 621 nm, and emit at 515 nm or above 650 nm. Alternatively, we accept biotin-adducts of your ligand that can easily be coupled to the desired fluorophores.
- They should be chemically stable and preferably not cytotoxic at test concentrations.
- They can target several GPCRs with similar affinities, their selectivity against other GPCRs is not a key criterion for the selection of the ligand (a broad coverage across sub-families would even be preferred).
- Optional: For planned assay validations, it will be beneficial to receive also unlabeled tracer control(s) as part of your submission.
For all submissions: The chemical structures of all submitted tracer(s) and control(s) SHOULD NOT be disclosed in your application. Only information on their molecular weight and spectral properties must be shared as part of this opn2EXPERTS call.
What potential solutions would be out of scope?
The following ligands will be considered out of scope:
- Binding affinities of the ligand far above 300 nM.
- Fluorescent moieties that don’t meet the spectral property criteria mentioned above.
- Chemically unstable and strong cytotoxic molecules (EC50 > 10 µM).
- Radioisotope labeled molecules.
- Commercially available molecules.
- Alternative technological solutions like e.g., SPR, DSF and others.
How is this opn2EXPERTS question structured?
This opn2EXPERTS call is structured in two main Phases and with the current call on opnMe.com, the first Phase is initiated. For detailed information please click on the respective chapters below.
The table below provides an overview on the different Phases of this call. The first Phase (Phase 1a) has been initiated and continues until September 27, 2023, 11:59 pm PST.
|
Phase |
|||||
|
Timeline |
June 11, 2023 – |
September 28, 2023 – |
April 1, 2024 – |
May 2, 2024 – |
April 1, 2025 – |
|
Our action |
This call is open for proposals. |
Characterization of your tracers:
|
April 1, 2024: Contacting the winners. |
Generation of GPCR selectivity profiling data on selected GPCR drugs. |
Publication |
|
Your action |
You send in GPCR tracers meeting criteria using submission form. |
|
You provide more of your tracer if required. |
For selected tracer molecules: Disclose your structures to us and give permission to include them in the final publication due December 31, 2025. |
|
|
IP |
IP remains with you. Don’t disclose structures. |
Your window to decide to patent your IP. |
IP strategy must be completed. |
||
|
Milestone |
All submissions comprise the final set of GPCR tracer candidates. |
Assay development complete. |
March 29, 2024: Selection of winners. |
GPCR dataset complete. |
Publication complete. |
|
Your benefits |
Get reimbursed for shipment (200 euros). |
Get characterization data for your tracer molecule including an optimized assay protocol. |
|
Get an aliquot of the TR-FRET Plasmid used for your tracer on demand. Selected tracer molecules win 2,000 euros. Submit small molecule test candidates to be considered for GPCR profiling. Get pre-publication access to the GPCR dataset. |
Co-authorship for selected final tracer. |
Phase 1 (June 12, 2023 – March 29, 2024)
Covers the reagents submission (Phase 1a) and assay development (Phase 1b) phases.
How to participate - Your action:
Until the end of the submission time of this first Phase (Phase 1a), we invite all experts in the field of GPCRs to share 1 mg of their fluorescent tracer(s) with us for the above-described receptors using the submission document provided with this call. To facilitate the pre-selection process, we highly recommend including data showing that your probes fulfill the above-mentioned criteria. The submission document will also require your signature as it serves as a material transfer agreement that defines and secures your intellectual property contribution that ONLY provides Boehringer Ingelheim with the license to develop and perform an assay based on your shared fluorescent probe (tracer). In fact, we kindly ask you not to disclose your fluorescent probe’s structures or SMILES codes at this point. Submissions are accepted through September 27th, 2023, 11:59 pm PST.
Our commitment:
On our end, we are committed to characterize the equilibrium binding and kinetic properties and the selectivities of all submitted tracers that meet the criteria listed above within six months (Phase 1b), and to share with each applicant all results that Boehringer Ingelheim generated, including a license to use these data freely and the right to publish after finalization of Phase 2.
Your benefits:
As a participant to this opn2EXPERTS call, you will have the chance to outline and share your expertise, skill set, and prior successes in the field with a top 20 pharmaceutical company. For Phase 1, you will receive 200 euros as a reimbursement of your shipping costs for the fluorescent tracer(s) delivery for a maximum of one delivery. We kindly ask you to refrain from sending individual shipments for individual tracers; rather we encourage you to combine the shipment. The assay development at Boehringer Ingelheim including the other reagents needed will be covered by Boehringer Ingelheim. You will receive all data that Boehringer Ingelheim will generate, and the prospect for a publication on this assay development. Overall, it opens the opportunity to work on a scientific topic in the field of GPCR and accelerate the drug development of potential compounds targeting essential GPCRs. All submitted IP will continue to stay with you and you are free to use it for other purposes.
Milestone:
Based on the data generated for all submitted tracer molecules, we will decide on a final tracer set that serves as the reagent collection for the future GPCR selectivity panel. As decision criteria for the selection, we apply our “in scope” criteria to move to Phase 2. To summarize, your tracer
- was detectable using the TR-FRET assay set-up,
- shows sufficient potency,
- shows favorable (fast) kinetic binding properties,
- shows no cytotoxicity at test concentrations, and
- was used to determine a control compound EC50 (for example using an unlabeled version of your molecule or a known drug) [optional]
to proceed to phase 2.
In addition, the number of submitted tracer candidates submitted per GPCR and person must be limited to 10 for logistical reasons. In case of similar entries from different participants that meet all quality criteria, we reserve the right to select the final molecule.
Phase 2 (April 1, 2024 – December 31, 2025)
Covers the winner selection (Phase 2a), GPCR selectivity assessment (Phase 2b), and publication (Phase 2c) phases.
Our commitment:
As part of Phase 2, we plan to characterize and study the selectivity of a broader small molecule GPCR drug set (> 300 drugs) both looking at equilibrium and binding kinetic properties where feasible based on the tracers submitted. After thorough analysis we plan to publish the results in a co-authored publication by December 31, 2025.
Depending on the outcome of in-depths characterization analyses conducted on our end as part of Phase 1, we might be interested in obtaining an additional sample of your submitted tracer(s). Should your tracer(s) be affected, we will contact you at that time for next steps.
Your benefits:
You will benefit from receiving a 2,000 euros award per accepted and validated fluorescent probe that will serve as a non-exclusive license fee. Also, you will receive a detailed protocol describing the assay conditions including detailed validation data. You may have the opportunity to receive the plasmid-DNA to set up the assay in your own laboratory. In addition, we offer you the possibility to co-author a broader scope scientific publication describing our overall efforts to improve early drug safety screening in a high impact factor journal upon conclusion of this call. To be eligible for co-authorship you should be willing to disclose your tracer structure in the publication.
Furthermore, you will have the opportunity to nominate a maximum of five compounds for validation studies and gain pre-publication access to the data of the broader GPCR panel generated. Your contribution will be valued in the community and offers you the possibility of high visibility as well as a network opportunity and link with likeminded scientists.
Key criteria for the best answer:
Your tracer(s) will be selected from all submissions based on the criteria listed in Phase 1. We reserve the right to select the winner based on the tracer properties including potency, binding kinetic properties, cytotoxicity, and the ability to be used for a successful EC50 determination. In case of multiple potential tracer submissions for one of the 66 GPCR assays, we may select the tracer that covers more than one GPCR to minimize reagents needed for the final panel.
What information should be included in your answer submission?
Please use our answer submission template to provide a 2-3 page non-confidential proposal (available for download here).
If confidential data exists that would strengthen the proposal, please indicate that information is available to share under a Confidential Disclosure Agreement (CDA). If we find the non-confidential concept proposal sufficiently interesting, we will execute a CDA for confidential discussions.
Summary
Take the opportunity to join our new opnMe GPCR Route 66+ call: Which of your fluorescent ligands can shed light on the selectivity of GPCR-targeting compounds to support faster drug discovery efforts?
All incoming tracer submissions will be evaluated by a scientific jury, and, upon selection, will be included in the opnMe GPCR Route 66+ assay panel. Submitting scientists will initially receive 200 euros reimbursement for their shipments (limited to one reimbursement per submitting scientist) and will receive exclusive access to all data generated with their tracer(s). Upon selection to the final assay panel, scientists whose tracer(s) will be selected will receive 2,000 euros as an award for each accepted and validated tracer. We will provide contributors of the final tracer set with the opportunity to propose candidate molecules for inclusion in the tests on the resulting broad GPCR panel to determine their affinities and kinetic binding properties. In addition, scientists will have the opportunity to co-author an open-access publication that summarizes the efforts of this call.
We can only accept tracer proposals if they arrive by the submission deadline on September 27, 2023, 11.59 pm PST.
References
Appendix – list of GPCRs included in the opnMe GPCR Route 66+ project
Download the full list of the 66 GPCRs included in the opnMe "Illuminating the GPCRome" call.
|
Uniprot Name |
Uniprot ID |
Name |
Ligand |
|
5HT1A_HUMAN |
P08908 |
Serotonin 5HT1A |
5-Hydroxytryptamine |
|
5HT1B_HUMAN |
P28222 |
Serotonin 5HT1B |
5-Hydroxytryptamine |
|
5HT1D_HUMAN |
P28221 |
Serotonin 5HT1D |
5-Hydroxytryptamine |
|
5HT1F_HUMAN |
P30939 |
Serotonin 5HT1F |
5-Hydroxytryptamine |
|
5HT2A_HUMAN |
P28223 |
Serotonin 5HT2A |
5-Hydroxytryptamine |
|
5HT2B_HUMAN |
P41595 |
Serotonin 5HT2B |
5-Hydroxytryptamine |
|
5HT2C_HUMAN |
P28335 |
Serotonin 5HT2C |
5-Hydroxytryptamine |
|
5HT4R_HUMAN |
Q13639 |
Serotonin 5HT4 |
5-Hydroxytryptamine |
|
5HT7R_HUMAN |
P34969 |
Serotonin 5HT7 |
5-Hydroxytryptamine |
|
ACM1_HUMAN |
P11229 |
Muscarinic acetylcholine receptor M1 |
Acetylcholine |
|
ACM2_HUMAN |
P08172 |
Muscarinic acetylcholine receptor M2 |
Acetylcholine |
|
ACM3_HUMAN |
P20309 |
Muscarinic acetylcholine receptor M3 |
Acetylcholine |
|
ACM4_HUMAN |
P08173 |
Muscarinic acetylcholine receptor M4 |
Acetylcholine |
|
ACM5_HUMAN |
P08912 |
Muscarinic acetylcholine receptor M5 |
Acetylcholine |
|
AA1R_HUMAN |
P30542 |
Adenosine A1 receptor |
Adenosine |
|
AA2AR_HUMAN |
P29274 |
Adenosine A2A receptor |
Adenosine |
|
AA2BR_HUMAN |
P29275 |
Adenosine A2B receptor |
Adenosine |
|
AA3R_HUMAN |
P0DMS8 |
Adenosine A3 receptor |
Adenosine |
|
ADA1A_HUMAN |
P35348 |
Alpha-1A adrenergic receptor |
Adrenaline |
|
ADA1B_HUMAN |
P35368 |
Alpha-1B adrenergic receptor |
Adrenaline |
|
ADA1D_HUMAN |
P25100 |
Alpha-1D adrenergic receptor |
Adrenaline |
|
ADA2A_HUMAN |
P08913 |
Alpha-2A adrenergic receptor |
Adrenaline |
|
ADA2B_HUMAN |
P18089 |
Alpha-2B adrenergic receptor |
Adrenaline |
|
ADA2C_HUMAN |
P18825 |
Alpha-2C adrenergic receptor |
Adrenaline |
|
ADRB1_HUMAN |
P08588 |
Beta-1 adrenergic receptor |
Adrenaline |
|
ADRB2_HUMAN |
P07550 |
Beta-2 adrenergic receptor |
Adrenaline |
|
ADRB3_HUMAN |
P13945 |
Beta-3 adrenergic receptor |
Adrenaline |
|
AGTR1_HUMAN |
P30556 |
Angiotensin receptor AT1 receptor |
Angiotensin |
|
CCR5_HUMAN |
P51681 |
C-C chemokine receptor type 5 |
Chemokine |
|
CXCR4_HUMAN |
P61073 |
C-X-C chemokine receptor type 4 |
Chemokine |
|
C5AR1_HUMAN |
P21730 |
C5a Receptor |
Complement peptide |
|
DRD1_HUMAN |
P21728 |
Dopamine D1 receptor |
Dopamine |
|
DRD2_HUMAN |
P14416 |
Dopamine D2 receptor |
Dopamine |
|
DRD3_HUMAN |
P35462 |
Dopamine D3 receptor |
Dopamine |
|
DRD4_HUMAN |
P21917 |
Dopamine D4 receptor |
Dopamine |
|
DRD5_HUMAN |
P21918 |
Dopamine D5 receptor |
Dopamine |
|
EDNRA_HUMAN |
P25101 |
Endothelin A receptor |
Endothelin |
|
EDNRB_HUMAN |
P24530 |
Endothelin B receptor |
Endothelin |
|
HRH1_HUMAN |
P35367 |
Histamine H1 receptor |
Histamine |
|
HRH2_HUMAN |
P25021 |
Histamine H2 receptor |
Histamine |
|
HRH3_HUMAN |
Q9Y5N1 |
Histamine H3 receptor |
Histamine |
|
HCAR2_HUMAN |
Q8TDS4 |
Hydroxycarboxylic acid receptor 2 |
Hydroxycarboxylic acid |
|
HCAR3_HUMAN |
P49019 |
Hydroxycarboxylic acid receptor 3 |
Hydroxycarboxylic acid |
|
CLTR1_HUMAN |
Q9Y271 |
Cysteinyl leukotriene receptor 1 |
Leukotriene |
|
CLTR2_HUMAN |
Q9NS75 |
Cysteinyl leukotriene receptor 2 |
Leukotriene |
|
MTR1A_HUMAN |
P48039 |
Melatonin receptor type 1A |
Melatonin |
|
MTR1B_HUMAN |
P49286 |
Melatonin receptor type 1B |
Melatonin |
|
NPY2R_HUMAN |
P49146 |
Neuropeptide Y receptor type 2 |
Neuropeptide Y |
|
NTR2_HUMAN |
O95665 |
Neurotensin receptor type 2 |
Neurotensin |
|
OPRD_HUMAN |
P41143 |
Opioid receptors δ receptor |
Opioid |
|
OPRK_HUMAN |
P41145 |
Opioid receptors κ receptor |
Opioid |
|
OPRM_HUMAN |
P35372 |
Mu-type opioid receptor |
Opioid |
|
OXYR_HUMAN |
P30559 |
Oxytocin receptor |
Oxytocin |
|
P2RY2_HUMAN |
P41231 |
P2Y purinoceptor 2 |
P2Y |
|
P2Y12_HUMAN |
Q9H244 |
P2Y purinoceptor 12 |
P2Y |
|
PTAFR_HUMAN |
P25105 |
Platelet-activating factor receptor |
Platelet-activating factor |
|
PE2R1_HUMAN |
P34995 |
Prostaglandin E2 receptor EP1 subtype |
Prostaglandin |
|
PE2R2_HUMAN |
P43116 |
Prostaglandin E2 receptor EP2 subtype |
Prostaglandin |
|
PE2R3_HUMAN |
P43115 |
Prostaglandin E2 receptor EP3 subtype |
Prostaglandin |
|
PE2R4_HUMAN |
P35408 |
Prostaglandin E2 receptor EP4 subtype |
Prostaglandin |
|
PI2R_HUMAN |
P43119 |
prostaglandin I2 receptor |
Prostaglandin |
|
PF2R_HUMAN |
P43088 |
Prostaglandin F2-alpha receptor |
Prostaglandin |
|
PAR1_HUMAN |
P25116 |
Proteinase-activated receptors |
Proteinase activated |
|
S1PR1_HUMAN |
P21453 |
Sphingosine 1-phosphate receptor 1 |
S1P |
|
S1PR5_HUMAN |
Q9H228 |
Sphingosine 1-phosphate Receptor 5 |
S1P |
|
GPR35_HUMAN |
Q9HC97 |
Kynurenic acid receptor |
Kynurenic Acid |